C. Progress Report
C.1. Previous aims and summary of progress
This report covers the period 8/1/2002-3/6/2007. We refer to this time interval as the current grant period. The specific aims during this period were: 1) develop a spatio-temporal experiment management system (STEMS) that handles complex multi-resolution brain map data, 2) develop methods for both humans and computers to interact with the database, including 3-D visualization of spatio-temporal results, and 3) develop methods for peer-to-peer sharing of independently maintained spatio-temporal experiment management systems
One of our primary accomplishments during this period was to formulate the vision and requirements we describe in the background (section B.3). This vision is a refinement of the vision we have had throughout, but our experience in the current and previous funding periods has clarified it. In terms of these requirements, we have made significant progress in local spatio-temporal data management, interoperability, methods for accessing local data, and methods for combining multiple data sources. This progress has led to a demonstration of the feasibility of our lightweight approach to data integration, as well as 6 software toolkits that are available for download, 11 online demos, a neuroanatomical ontology, and a database of over 100 cortical stimulation mapping (CSM) patients [1]. These tools and data have been used to demonstrate dissociation between CSM sites for actions and those for object naming, various correlation studies among multimodality language data, and studies of face processing in individuals with autism.
In the following sections we first describe the data that have provided the driving biological problem for our tool development, after which we describe in more detail the specific progress in meeting the requirements, and the impact that this progress has had in terms of neuroscience results and outside use of and interest in our tools. Although the progress is organized according to requirements needed to achieve our proposed vision, we indicate in parentheses the specific aims from the current grant period that each accomplishment addresses.
C.2. Visualization based mapping protocols
Our work to-date has been largely driven by the data management and integration needs of Drs. George Ojemann and David Corina. Dr. Ojemann is a neurosurgeon who has published extensively on the use and integration of neurosurgical data for understanding language organization in the brain [2] [3] [4-11]. One such type of data is that obtained from Cortical Stimulation Mapping (CSM), which is a procedure routinely done during resection of temporal lobe epileptic foci in order to avoid areas of language cortex [5].
In this procedure a series of numbered tags marking sites for stimulation (CSM sites) are placed at various locations on the exposed cortical surface, and an intraoperative photograph is taken of the tags. The patient is then awakened, and a small electrical current is applied to the stimulation sites while the patient is asked to perform various language tasks. If the patient is unable to perform the language tasks while the site is stimulated that site is avoided during the resection since studies have shown that disruption can lead to aphasia [2].
In our previously funded work we developed a software tool, called the Visual Brain Mapper (VBM), for mapping these CSM sites to a 3-D patient-specific model so that they could be related to other types of data in order to gain a better understanding of language function. VBM, which was fully documented during the current grant period [12], takes as input the intraoperative photograph and a set of structural MRI volumes acquired approximately one week prior to surgery by Dr. Corina. It produces as output a 3-D model of the patient’s own brain, together with the 3-D patient-specific coordinates of the CSM sites mapped to the 3-D model. The mapping permits the CSM sites to be related to other multimodality data, such as fMRI and Single Unit Recording (SUR) that are acquired and registered to the same patient-specific MR-based coordinate system.
The fMRI data are acquired during the MR imaging session prior to surgery, and analyzed and registered to the structural MRI using SPM [13]. SUR data are acquired at the time of CSM mapping. In this case a tungsten electrode is inserted into an area of cortex to be resected, the location of which is marked by a numbered tag, and subsequently mapped to 3-D patient coordinates using the VBM procedure. The electrode records the response of neurons close to the buried electrode tip. At a later time individual neurons can be separated by a spike sorting procedure we developed during the current grant period [14], and the firing rates and patterns of the neurons in response to stimuli can be compared to co-registered fMRI or CSM data.
C.3. Local spatio-temporal data management
C.3.1. STEMS toolkits (previous aim 1)
To manage these data and make them accessible for integration we created several STEMS, which we now refer to as Laboratory Information Management Systems (LIMS) because the acronym “LIMS” is in more widespread use. During the current grant period we created several LIMS toolkits, and used them to create separate LIMS for managing three different types of Ojemann/Corina data: CSM, fMRI and SUR. We also developed and analyzed a survey, which demonstrated that many other researchers need these kinds of tools [15].
The Web Interfacing Repository Manager (WIRM) [16] consists of a set of Perl libraries that allow a programmer to create a web-based front-end to a backend relational database, and to manage uploaded image and other multimodality data through metadata stored in the database. WIRM is open source, and has been used in several database management projects developed by us [17, 18] or a spinoff company [19]. An extension of WIRM is the Customized Electronic Laboratory Online (CELO), which is designed to allow a non-programmer to create a relatively simple LIMS through a web interface [20]. In other funded work we have used CELO to create a LIMS for a lab studying lens cataracts [21] and a prototype for a proteomics lab studying neurodegenerative disease [22]. We are also involved in a pilot project to evaluate CELO and Seedpod (described below) for their utility in creating LIMS for clinical trials management.
The initial driving application for the creation of WIRM was management of the CSM data, with the result that our WIRM-based CSM database [18] is the most mature of our local LIMS. The CSM LIMS currently includes 87 surgical patients. It is in active use by us and our collaborators for patient demographics, workflow management for VBM, recording of CSM maps generated by VBM, and post-surgical coding of CSM language errors. In the latter case a psychology student listens to audio tapes of patient responses to cortical stimulation, and classifies the responses according to a coding scheme that is embedded in the database [23]. Example error codes are “Semantic paraphasia”, which is related to meaning, and “Phonological paraphasia”, which is related to syntax. The data saved in the CSM database (Figure 1A) are used in many of the applications described below.
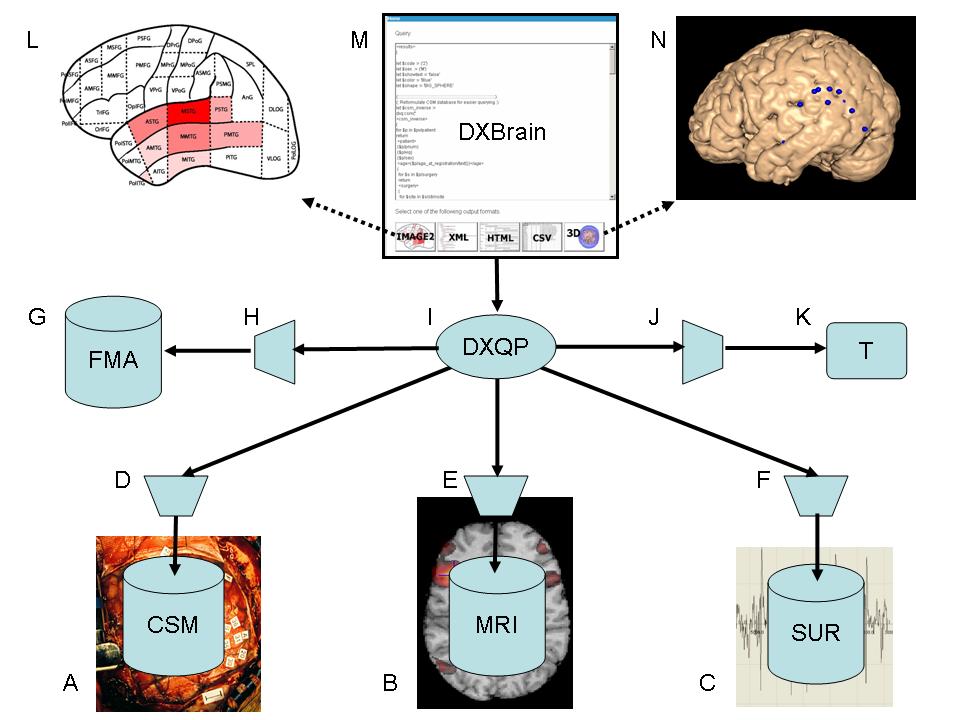
Figure 1 DXBrain local data integration network
Another application is called XBatch, [24, 25] which was initially developed to manage the Ojemann/Corina MRI data. XBatch transparently embeds data management behind a batch processing toolbox within the SPM fMRI analysis package [13]. XBatch permits the user to generate batch scripts that run complex fMRI analyses without user intervention, while at the same time recording these operations, as well as the locations of the input and output files, as instances of classes in a Protégé [26] ontology that is based on the Dartmouth fMRI Data Center experiment lab book ontology [27]. At any time this ontology can be examined in Protégé. We have also written scripts to transform the Protégé ontology into an XML database that can be searched using XQuery, the query language for XML [28]. Within SIG we have created two such XML fMRI databases using XBatch: one for the Ojemann/Corina fMRI data, and one for researchers using fMRI to study face processing in autism (section B.1). Each is encapsulated as a web service searchable by XQuery (section C.5.2), and each is one of the data sources available for search by our distributed data integration application (Figure 1B).
A more ambitious LIMS toolkit is the Seedpod model-driven approach to LIMS creation [29, 30]. In this case the developer uses Protégé to create an ontology describing the objects and relations representing the data to be managed. The ontology not only describes the data, it also describes the expected behavior of a web-based data management system, including the types of widgets that will be used to enter and display data. Given this ontology the developer runs a program that transforms the Protégé ontology into a relational database schema, which includes not only standard tables describing the entities and their relations, but also metadata tables describing the system behavior. The schema is loaded into a PostgreSQL [31] database, after which a Java-based web application interprets the schema to generate a web front-end. The driving application for the development of Seedpod is management of SUR data (Figure 1C). Seedpod is still under development as an informatics PhD thesis.
C.3.2. Content-based retrieval (aim 1)
The LIMS described in section C.3.1 manage multimodality data by maintaining metadata in a backend relational database that describes, among other things, the file locations of images and other uploaded files.
We explored more precise spatio-temporal queries of these data through the development of a separate MatLab-based content-based retrieval system, using fMRI, SUR and CSM data saved in the LIMS [32].
The system supports queries of the type, “Retrieve all fMRI activations that are close to the location of a given SUR recording in normalized space”, or “retrieve all SUR recordings that exhibit a firing pattern similar to a given SUR recording”. To support these queries we developed methods for characterizing SUR firing patterns, not only by the typical average firing rate, but also by the similarity of a given recording to a set of 16 firing patterns derived from a training set (8 patterns in the frequency domain and 8 in the time domain). These similarities were used to create a set of indices for rapid retrieval of the raw data files of SUR and other signals that were most similar to each pattern. This work is being continued as part of, and in fact formed much of the preliminary results for an NSF grant entitled, “Multimedia Information Retrieval for Biological Research” which is led by our co-investigator Linda Shapiro in Computer Science. In future work we will incorporate the results of the NSF work in the XML-based data integration system to be developed in the proposed research.
C.3.3. Visualization (previous aim 2)
Server-based visualization. The data files generated by many of the procedures described in section C.2 are saved on our Structural Informatics Group (SIG) servers, where they are made available to the VBM program (section C.2) through the CSM and fMRI databases (section C.3.1). In addition to its use in standalone mode, VBM can run in server mode, in which case the server accepts commands to load the 3-D models, fMRI and CSM data, render an image of the 3-D scene, send the image to a client application (generally a Java applet), and respond to commands to change the viewpoint or display different data [33]. This mode is in routine use by the neurosurgeons, who use the locations of fMRI activations to plan the sites of SUR as part of a study to correlate SUR activity with fMRI (section C.7).
MindSeer. The VBM program, while very useful, is not portable or particularly interactive. Thus, in the current grant period we implemented a Java3D-based visualization and mapping program, called MindSeer, that replicates the features of VBM, runs on all major platforms (Linux, Windows, Mac), runs at interactive speeds, runs in both stand-alone and client-server mode, and is extensible through the use of plugins [34-36]. Currently, MindSeer fully supports the data types and file formats used by SPM [13], and additional image, model and visualization formats can be easily added through the plugin mechanism [37]. Current visualization modes include the standard orthogonal volume slice view and 3-D surface view with cutaway. FMRI, CSM and other functional data may be displayed on slice or 3-D surface views.
MindSeer organizes data through an XML-based I/O specification file called the “library”, which allows the user to easily access all data needed for visualization. In our data integration work (section C.6) we dynamically generate this library to show only the results of database queries. A poster presentation of MindSeer received best poster out of over 400 posters at the premier international medical informatics meeting [34], and a journal article has been submitted [36].
C.4. Local Interoperability
C.4.1. Symbolic Interoperability (previous aim 1)
Our primary method for achieving symbolic interoperability is in terms of the neuroanatomical extensions we added to our Foundational Model of Anatomy ontology during the current grant period, and the methods we developed for accessing the FMA.
The Foundational Model of Anatomy (FMA) ontology (Figure 1G) is our symbolic representation for the structure of the body, and our primary proposed framework for organizing other biomedical information [38-41] [42]. The ontology is implemented Protégé, is stored in a relational database, and currently contains 75,000 classes and 2 million relationships representing structures ranging in size from biological macromolecules to the whole body. The detail and scope of represented anatomical knowledge far exceed any currently available computable anatomical resources.
During the current grant period we initiated the development of the neuroanatomical component of the FMA (the Neuroanatomical FMA, or NFMA) by adding NeuroNames [43] terminologies and relationships in their proper locations in the larger FMA ontology [44, 45]. The long term goal of this work is to use the NFMA as a basis for reconciling many of the conflicting schemes for representing the anatomy of the brain. In the current grant period we developed methods to utilize the current version of the NFMA in data integration by first creating a parcellated schematic view of the left side of the cortex, and labeling each of the parcels with names from the NFMA [46] (Figure 1L). This parcellation was then used by a neuroanatomist to annotate each mapped CSM site with a name from the NFMA. The neuroanatomist did this by visually identifying the brain parcel on the 3-D rendering of the patient brain surface (section C.2) that contained the given CSM site. The resulting terms are stored in the CSM database, thereby allowing CSM sites from different patients to be coarsely integrated through anatomical names.
To facilitate access to the FMA we implemented a query-based interface called OQAFMA (OQAFMA Querying Agent for the Foundational Model of Anatomy) [47]. OQAFMA takes as input a query expressed in the StruQL query language, which is optimized for large graphs, and was co-developed by our co-investigator Dan Suciu [48]. The StruQL query is reformulated as a set of SQL statements to an optimized (for efficiency) version of the relational database that stores the FMA. Results are returned as XML. A web service wrapped version of OQAFMA is one of the sources accessible from our distributed data integration application (Figure 1H).
OQAFMA, the FMA and the query reformulation methods described below (section C.5.1) were the basis for a recently-funded BISTI collaborative grant, led by the PI, with the Stanford Center for Biomedical Ontology, which will develop a query-based framework for deriving application ontologies from reference ontologies as part of the fractal semantic web [49]. Eventually we envision fractal data management networks, of the type we propose in this grant, interacting with the fractal semantic web, in both cases through query interfaces.
C.4.2. Spatial interoperability (previous aim 1)
Annotating each CSM site with an FMA term provides a coarse form of interoperability of the type that is commonly communicated in the neuroscience literature (e.g. “activation occurred in the middle temporal gyrus”). More precise interoperability requires methods for spatial normalization (section B.4). In the current grant period we developed a methodology for choosing which of the available methods would be of most use, under the assumption that the most useful method would vary according to the problem, which in our case is normalization of CSM sites [50-52].
In collaboration with the David Van Essen lab we applied this methodology to CARET [53] surface normalizations and SPM [13] volume normalizations for 11 subjects from our CSM database. We found that there was no significant difference between the two methods, and that SPM was faster and easier to use. Based on this analysis we used SPM to perform volume-based non-linear spatial registration of 72 (out of a total of 79) structural MRI volumes in our CSM database to the MNI normalized space used in SPM. Seven could not be normalized because of distorted brain anatomy, generally due to tumor or previous resections. We saved the resulting deformation field for each patient, and made each one accessible to a transformation web service that, when given a patient number and a set of magnet coordinates for a CSM site, returns the normalized coordinates. This “spatial normalization” service T (Figure 1K) then permits the mapping of CSM sites from multiple patients onto any structural MR surface or volume that has been transformed to MNI space. The transformation web service is another web service encapsulated source that is accessible from our distributed data integration application (Figure 1J).
C.5. Methods for accessing local data
C.5.1. XML query interface to relational data (previous aim 2)
The LIMS described in section C.3.1 permit drill-down navigation of the data through a web interface, but they do not permit ad hoc queries needed for data mining and sharing. Therefore, we implemented a method for dynamically publishing relational data as XML, since XML has become the medium of data exchange.
The implemented method, which is called SilkRoute II [54, 55] is a second generation version of a middleware program, called SilkRoute, developed by Dan Suciu and others [56]. Like the first version (which was never fully operational), SilkRoute II [54, 55] defines a “canonical” XML view of a relational schema, in which each row in a relational table is an XML element, and the columns of the table are sub-elements. The developer writes an XQuery over the canonical view, which defines an XML “public view” over the relational database. Unlike the canonical view (which is the XML view currently provided by some relational database vendors), the public view can create an arbitrary XML tree out of elements in the canonical view, and can rename or omit elements, thus better matching the structure of the data as perceived by the user. Given the public view the user writes an arbitrary XQuery against it. The public view looks to the user as though it is an XML document, whereas in fact it is a non-materialized view defined by an XQuery. The SilkRoute engine composes the user XQuery against the XQuery defining the public view, emitting a series of SQL commands to the underlying relational database. The returned results from the database are reformulated as XML, appearing to the user as though the user XQuery had been directly processed against the public view materialized as an XML document.
The advantages of this approach for our purposes are 1) the backend relational database can remain unchanged, without requiring expensive porting to an XML database, and 2) the public view acts as a filter that allows the developer to only present data that is desired to be shared.
We implemented SilkRoute II as an XML “wrapper” over the CSM database, and developed a web application, called XBrain, for querying the CSM database through the SilkRoute II wrapper [57, 58]. However, further use of SilkRoute II revealed that is not yet fully compliant with the full XQuery specification, and it can have very slow response time (on the order of minutes). Therefore, for our data integration work we implemented a daily cron job that calls SilkRoute II to materialize the public view of the CSM database as a large XML file, then encapsulates an XQuery interface to the materialized view as yet another web service (section C.5.2) accessible to our data integration application (Figure 1D). The materialized view takes about 7 seconds to generate, and is 25 megabytes in size. However, this approach will not scale since many data sources are much larger than our CSM database. We will address this issue in aim 1 of the proposed work.
C.5.2. WIX
The XQuery web service interface to an XML database or materialized XML view of a relational database is generated by a small Java tool we created called WIX (Web Interface to XML) [59]. This tool is an executable Jar (java archive) file that presents an interface allowing a user to specify an XML file, an optional XML schema for the XML file, and a name for the web service. The tool then generates a War (web archive) file that can be deployed to an Apache Tomcat server [60] (or other J2EE container) by simply dropping the war file in the proper location. The XML file then becomes accessible for query by XQuery via the encapsulated web service. The advantage of this approach over the standard “document” directive in XQuery is that only the query is shipped over the network, not the entire, possibly large document. Wix uses the Saxon XQuery processor [61] as the underlying XQuery engine. Wix enabled XQuery web-service interfaces to data are represented in Figure 1D, E, and F.
C.6. Local distributed query processing (previous aim 3)
Given the existence of separate LIMS for each data type (section C.3.1), symbolic and spatial interoperability methods (sections C.4.1 and C.4.2), methods for querying the databases (section C.5) and methods for visualizing the results (section C.3.3), the next problem is how to query over all these separate databases and view the integrated results.
We investigated several approaches to this problem in the current grant period: 1) peer to peer integration similar to the original Napster music sharing program , and based on the Piazza system being developed in UW CS [62], 2) mediator-based integration based on the UW Biomediator project [63], and 3) distributed XML query processing. We found that, for the small scale, mostly non-overlapping data sources available from our primary driving biological projects, the peer to peer approach was more difficult than either we or our UW database collaborators had anticipated, and the (current) mediator-based approach was too slow and “heavyweight”, requiring significant informatics effort to maintain a centralized mediated schema [64, 65].
Thus, as noted in section B.3 we have chosen to pursue the third approach, which we call lightweight distributed queries [66]. The current instantiation of this architecture is shown in Figure 1. At the bottom outer level are a series of source databases, services or ontologies, each wrapped as a web service (indicated by trapezoids, Figure 1D,E,F,H,J), and all returning XML. As described in the previous sections, the web services we have implemented to-date are 1) the FMA via OQAFMA, 2) the CSM database via materialized XML public views generated by SilkRoute II, 3) the fMRI database created by XBatch and converted from Protégé to an XML document, 4) an XML snapshot of the SUR database, and 5) the transformation service T, which transforms patient-specific coordinates to normalized coordinates. The CSM and fMRI databases include the locations of multi-modality files.
C.6.1. DXQP
The central integrator in our system is another Java tool called DXQP, or Distributed XQuery Processor, shown in Figure 1I [67]. DXQP, like Wix (section C.5.2) is an extension of the Java Saxon XML Query processor [68]. The extension exports an XQuery function, called dxq:xquery, which can call any web service that returns XML. Calls to this function, with different web services and different XQueries or other commands to these services, can then be embedded within a larger XQuery, thus allowing the distribution of a query over multiple sources.
C.6.2. DXBrain
DXQP is in turn accessed by a web front-end called DXBrain (Distributed XBrain, Figure 1M), which is an extension to the XBrain system (section C.5.1) that demonstrates the use of distributed XQueries over the separate web service encapsulated sources shown in the figure. The user accesses DXBrain through a web address which is linked to from our demo page [1]. Public users are only able to see a subset of the patients, but logged in users with the proper permissions can see all the data. The user can create a new XQuery or select a previously saved XQuery and modify it as needed. The top center of Figure 1 represents an example XQuery over the CSM database that requests all CSM sites located in the temporal lobe, which were involved in a CSM trial in which the patient made a language error coded as type 2, which is semantic paraphasia (section C.3.1). Once the XQuery has been formulated the user clicks one of the buttons at the bottom (of the top central screenshot) to submit the query to DXQP, where it is sent to the individual sources. The XML results returned from the sources are combined as a single XML result. For the example query the result includes each site in the temporal lobe with an error code of type 2, and for each of these sites, the FMA name associated with that site, as well as the patient-specific 3-D magnet coordinates of the site as defined by the VBM procedure (section C.2). Depending on which button was clicked the raw results are transformed by XLST for viewing as XML, HTML, or CSV (for input to Excel). In addition, if the user clicks the Image2 or 3-D buttons the results are presented as dynamically-generated brain-specific images.
For Image2 the results are mapped to the 2-D parcellated brain schematic described in section C.4.1, in which case an image is dynamically generated that colors each parcel according to how many sites with language error 2 are retrieved for that parcel (Figure 1L). If the user clicks the 3-D button the system first contacts the transformation service T to transform the patient-specific coordinates for each site to normalized coordinates. It then writes a MindSeer library file (section C.3.3) with the transformed sites, and creates a Java Web Start link to call MindSeer with the generated library file. When the user clicks the link MindSeer is invoked on the user’s workstation with the normalized query results preloaded. The user may then select a 3-D surface on which to display the sites. Current surfaces are the MNI average brain and the Collin27 atlas. The resulting display shows the 3-D locations of all sites that contained any trial with semantic paraphasia in the temporal lobe (Figure 1N). A journal article describing this system has been submitted [69].
C.6.3. Graphical XQuery generation (previous aim 2)
Recognizing that most neuroscience users will not want to learn the details of XQuery we have experimented with graphical methods for creating an XQuery over an XML document. The current implementation of this project, which is called XGI [70] uses the Ajax framework [71] to generate a dynamic web page. The application parses a schema describing the XML document and creates a collapsible tree (like MS Windows Explorer) that displays the elements of the schema on the left of the screen. The user selects elements from this tree and pastes them into a tree on the right side of the screen that becomes the structure of the constructed query. Constraints and predicates are added by typing them in a text box, and clicking to associate the constraints with the appropriate result element. Thus, this work does not try to completely specify the entire query using graphical methods, but mixes a text-based approach with graphical layout of the overall query structure. It also is only able to generate a query over a single source at a time, and does not permit the specification of all aspects of XQuery. We will build on this application in aim 2 of the proposed work (section D.2).
C.7. Neuroscience results
As noted earlier the Visualization Based Mapping and visualization tools, together with the data in the CSM database, are routinely used, by several geographically diverse collaborating labs, to relate CSM and SUR recording 3-D locations to fMRI and other functional imaging data. These tools have helped to provide suggestive evidence that there is no clear relationship between CSM and fMRI measures of language [72], an observation that has since been replicated elsewhere [73]. Further neuroscience research will be needed to better understand why these modalities are not correlated. However, the main point for the purposes of our proposed work is that these kinds of correlative studies, whether or not they show correlations, were greatly facilitated by the data management, integration and visualization tools we have developed.
Although there does not seem to be significant correlation (yet) among fMRI and CSM on the neurosurgical patients, the CSM data alone have still yielded significant results. In particular, analysis of these data have shown several patients with dissociation between language sites with action naming errors and those with object naming errors [74]. In addition, with the recent completion of the prototype XML-based query system coupled with 3-D visualization (section C.6), we expect to gain a more detailed understanding of the distribution of language processing as observed with CSM, than was found in the earlier Ojemann studies [5]. See letter of collaboration from David Corina. Thus, we will continue to “mine” the current CSM database as we create “views” of this unique data source that can be accessed by collaborating labs in aim 5.
C.8. Use outside the initial driving problem
Irrespective of whether correlations are found among the specific modalities and language tasks studied to-date by the Ojemann/Corina labs, there remains great interest in the wider neuroscience community in integrating multi-modality brain mapping data. In fact, several of our tools (XBatch, visualization applet, MindSeer) have been used by the local UW autism center to study face recognition in autism using EEG source localization and fMRI [75-77]; a project studying Transcranial Magnetic Stimulation (TMS) in the study of depression used our mapping tools to localize the position of the TMS probe; and we are in the process of applying our tools to a study of change in brain morphology in adult learners of a second language.
The operation of many of our tools can be seen in our demo page [1], and many are available for download from the same location. Although we have only recently begun tracking download statistics for many of these programs, the following table provides some indicator of their use by others:
|
Tool Name |
Number of downloads since Dec, 2006 |
|
FMA |
29 |
|
WIRM |
12 |
|
CELO |
6 |
|
XBatch |
211 (total downloads) |
|
MindSeer |
25 |
We have only recently made the WIX and DXQP programs available, so do not have download statistics for them. In addition we have received interest in these tools from several outside users. See letters from Gardner, Martone, Kikinis and Jacobs.
D. Literature Cited
1. Brinkley J. UW Integrated Brain Project: demos and downloads. 2006. http://sig.biostr.washington.edu/projects/brain/demos.html.
2. Ojemann GA. Brain organization for language from the perspective of electrical stimulation mapping. Behav Brain Sci 1983;6:189-206.
3. Ojemann GA. Electrical stimulation and the neurobiology of language. Behav Brain Sci 1983;6:221-226.
4. Ojemann GA, Dodrill CB. Verbal memory deficits after left temporal lobectomy for epilepsy: mechanism and intraoperative prediction. J. Neurosurg. 1985;62(1):101-107.
5. Ojemann G, Ojemann J, Lettich E, Berger M. Cortical language localization in left, dominant hemisphere: an electrical stimulation mapping investigation in 117 patients. J. Neurosurgery 1989;71:316-326.
6. Ojemann GA, Fried I, Lettich E. Electrocorticographic (ECoG) correlates of language. I. Desynchronization in temporal language cortex during object naming. EEG Clin Neurophysiol 1989;73:453-463.
7. Ojemann GA. Cortical organization of language and verbal memory based on intraoperative investigations. Progress in Sensory Physiology 1991;12:193-230.
8. Ojemann GA. Functional mapping of cortical language areas in adults, intraoperative approaches. In: Devinksy O, Berie A, Dogali M, editors. Electrical and magnetic stimulation of the brain and spinal cord. New York: Raven press. ltd.; 1993. p. 145-153.
9. Ojemann GA, Schoenfield-McNeill J. Neurons in human temporal cortex active with verbal associative learning. Brain and Language 1998;64:317-327.
10. Ojemann GA, Schoenfield-McNeill J. Activity of neurons in human temporal cortex during identification and memory for names and words. The Journal of Neuroscience 1999;19(13):5674-5682.
11. Ojemann GA, Schoenfield-McNeill, Lettich E, Oskin N, Zanos S, Panagiotides H, Zamora L, Sengupta C, Cho H, Corina D, Poliakov A, Martin RF, Brinkley JF. The relationship between fMR, single neuronal activity, and local field potentials in human association cortex. In: Proceedings, Society for Neuroscience Annual Meeting; 2005. p. 814.9.
12. Hinshaw KP, Poliakov AV, Martin RF, Moore EB, Shapiro LG, Brinkley JF. Shape-based cortical surface segmentation for visualization brain mapping. Neuroimage 2002;16(2):295-316 http://sigpubs.biostr.washington.edu/archive/00000047/.
13. Wellcome Department of Cognitive Neurology. Statistical Parametric Mapping. 2001. http://www.fil.ion.ucl.ac.uk/spm/.
14. Cho H, Corina D, Brinkley JF, Ojemann GA, Shapiro JG. A new template matching method using variance estimation for spike sorting. In: Proceedings, IEEE EMBS 2nd International Conference on Neural Engineering. ; 2005. http://sigpubs.biostr.washington.edu/archive/00000169/.
15. Anderson NR, Lee ES, Brockenbrough JS, Minie ME, Fuller S, Brinkley J, Tarczy-Hornoch P. Issues in biomedical data management and analysis: needs and barriers. Submitted. 2007.
16. Jakobovits RM, Rosse C, Brinkley JF. An open source toolkit for building biomedical web applications. J Am Med Ass. 2002;9(6):557-590 http://sigpubs.biostr.washington.edu/archive/00000134/.
17. Brinkley JF, Jakobovits R, Rosse C. An online image management system for anatomy teaching. In: Proc. AMIA Fall Symposium; 2002. p. 983. http://sigpubs.biostr.washington.edu/archive/00000120/.
18. Brinkley JF, Jakobovits RM, Poliakov AV, Martin RF, Gibson ER, Corina DM, Ojemann GA. An experiment management system for cortical stimulation mapping data. In: Society for Neuroscience Annual Meeting. San Diego; 2004. p. 1032.12. http://bmap.biostr.washington.edu/.
19. Vivalog. MyPacs.net. 2005. http://www.mypacs.net/.
20. Fong C, Brinkley J. Customizable Electronic Laboratory Online (CELO): A web-based data management system builder for biomedical research laboratories. In: Proceedings, Fall Symposium of the Amercian Medical Informatics Association; 2006. p. 922. http://sigpubs.biostr.washington.edu/archive/00000191/.
21. Fong C, Rosse C, Clark JI, Shapiro L, Brinkley JF. An ontology-based image repository for a biomedical research lab. In: MEDINFO 2004; 2004. p. 1598. http://sigpubs.biostr.washington.edu/archive/00000144/.
22. Fong C, Brinkley JF. Sample query from a prototype personal proteomics lab managment system. 2005. http://celo.biostr.washington.edu/celo-cgi/celo/make-sql.pl?ss_id=5&cx_ds_id=2&cx_lab=prota.
23. Corina D. Codes for CSM speech errors. 2006. http://bmap.biostr.washington.edu/repos/bmap_repo/Corinacoding.html.
24. Hertzenberg X, Poliakov A, Brinkley JF. Unobtrusive integration of data management with FMRI analysis. In: Proceedings, MEDINFO. San Francisco, CA.; 2004. p. 1639. http://sigpubs.biostr.washington.edu/archive/00000153/.
25. Poliakov A, Hertzenberrg X, Moore EB, Corina D, Ojemann GA, Brinkley JF. Unobtrusive integration of data management with fMRI analysis. Neuroinformatics. In Press 2006.
26. Gennari JH, Musen MA, Fergerson RW, Grosso WE, Crubezy M, Eriksson H, Noy NF, Tu SW. The evolution of Protege: an environment for knowledge-based systems development. Int. J. Human-Computer Studies 2003;58(1):89-123 http://www.smi.stanford.edu/pubs/SMI_Reports/SMI-2002-0943.pdf.
27. Van Horn JD, Grethe JS, Kostelec P, Woodward JB, Aslam JA, Rus D, Rockmore D, Gazzaniga MS. The Functional Magnetic Resonance Imaging Data Center (fMRIDC): the challenges and rewards of large-scale databasing of neuroimaging studies. Philos Trans R Soc Lond B Biol Sci 2001;356(1412):1323-1339.
28. Chamberlin D, Florescu D, Robie J, Simeon J, Stefanascu M. XQuery: A query language for XML. Boston: World Wide Web Consortium; 2001.http://www.w3.org/TR/xquery.
29. Li H, Brinkley JF, Gennari J. Semi-automatic database design for neuroscience experiment management systems. In: Proceedings, MEDINFO. San Francisco, CA.; 2004. p. 1639. http://sigpubs.biostr.washington.edu/archive/00000145/.
30. Li H, Gennari JH, Brinkley JF. Model driven laboratory information management systems. In: Proceedings, Fall Symposium of the American Medical Informatics Association; 2006. p. 484-488. http://sigpubs.biostr.washington.edu/archive/00000189/.
31. Stonebraker M, Kemnitz G. The POSTGRES next-generation database management system. Communications of the ACM 1991;34(10):78-92.
32. Cho H. Classification of functional brain data for multimedia retrieval [PhD Thesis]. Seattle: University of Washington; 2005.http://sigpubs.biostr.washington.edu/archive/00000186/
33. Poliakov AV, Albright E, Hinshaw KP, Corina DP, Ojemann G, Martin RF, Brinkley JF. Server-based approach to web visualization of integrated 3-D brain imaging data. Journal of the American Medical Informatics Association 2005;12(2):140-151 http://sigpubs.biostr.washington.edu/archive/00000170/.
34. Moore E, Poliakov A, Brinkley JF. Brain visualization in Java3D. In: Proceedings, MEDINFO. San Francisco, CA; 2004. p. 1761. http://sigpubs.biostr.washington.edu/archive/00000151/.
35. Moore EB, Poliakov A, Brinkley JF. Web-enabled 3D multimodality brain visualization in Java. In: Society for Neuroscience Annual Meeting; 2005. p. 570.2.
36. Moore EB, Poliakov A, Lincoln P, Brinkley J. MindSeer: A portable and extensible tool for visualization of structural and functional neuroimaging data. Submitted. 2007.
37. Lincoln P. Surface projection method for visualizing volumetric data. Seattle: University of Washington, Dept Computer Science and Engineering, Senior Thesis; 2006.http://sigpubs.biostr.washington.edu/archive/00000193/.
38. Brinkley JF, Eno K, Sundsten JW. Knowledge-based client-server approach to structural information retrieval: the Digital Anatomist Browser. Computer Methods and Programs in Biomedicine 1993;40:131-145.
39. Rosse C, Ben Said M, Eno KR, Brinkley JF. Enhancements of Anatomical Information in UMLS Knowledge Sources. In: Proceedings, 19th Annual Symposium on Computer Applications in Medical Care. New Orleans; 1995. p. 873-877.
40. Rosse C, Mejino JL, Modayur BR, Jakobovits RM, Hinshaw KP, Brinkley JF. Motivation and organizational principles for anatomical knowledge representation: the Digital Anatomist symbolic knowledge base. Journal of the American Medical Informatics Association 1998;5(1):17-40 http://sig.biostr.washington.edu/publications/online/KBpaper.pdf.
41. Rosse C, Shapiro LG, Brinkley JF. The Digital Anatomist foundational model: principles for defining and structuring its concept domain. In: Proceedings, American Medical Informatics Association Fall Symposium. Orlando, Florida; 1998. p. 820-824. http://sig.biostr.washington.edu/publications/online/D005094.pdf.
42. Rosse C, Mejino JLV. A reference ontology for bioinformatics: the Foundational Model of Anatomy. Journal of Bioinformatics. 2003;36(6):478-500 http://sigpubs.biostr.washington.edu/archive/00000135/.
43. Bowden DM, Dubach MF. An in-depth review of neuronames and brain info commentary. Neuroinformatics 2003;1(1):43-60.
44. Martin RF, Mejino JLV, Bowden DM, Brinkley JF, Rosse C. Foundational model of neuroanatomy: implications for the Human Brain Project. In: Proc AMIA Annu Fall Symp. Washington, DC; 2001. p. 438-442. http://sigpubs.biostr.washington.edu/archive/00000067/.
45. Martin RF, Mejino JLV, Bowden DM, Brinkley JF, Mulligan K, Rosse C. A next generation knowledge source: foundational model of neuroanatomy. In: Society for Neuroscience Annual Meeting. San Diego; 2001. p. 23.48.
46. Martin RF, Brinkley JF, Hertzenberg X, Poliakov A, Corina D, Ojemann GA. Anatomical parcellation of cortical language sites. In: MEDINFO; 2004. p. 1742. http://sigpubs.biostr.washington.edu/archive/00000156/.
47. Mork P, Brinkley JF, Rosse C. OQAFMA Querying Agent for the Foundational Model of Anatomy: a prototype for providing flexible and efficient access to large semantic networks. J. Biomedical Informatics 2003;36(6):501-517 http://sigpubs.biostr.washington.edu/archive/00000136/.
48. Fernandez M, Florescu D, Levy H, Suciu D. A query language for a web site management system. SIGMOD Record 1997;26(3):4-11.
49. Brinkley JF, Suciu D, Detwiler LT, Gennari JH, Rosse C. A framework for using reference ontologies as a foundation for the semantic web. In: Proceedings, Fall Conference of the American Medical Informatics Association; 2006. p. 96-100. http://sigpubs.biostr.washington.edu/archive/00000188/.
50. Smith VS. Evaluating Spatial Normalization Methods for the Human Brain [Masters Thesis]. Seattle: University of Washington; 2005.http://sigpubs.biostr.washington.edu/archive/00000173/
51. Smith VS, Hanlon D, Martin RF, Poliakov AV, Brinkley JF, Shapiro LS, Van Essen D, Ojemann GA, Corina DM. Evaluating anatomical normalization methods. In: Society for Neuroscience Annual Meeting. San Diego; 2004. p. 12.11.
52. Smith VS, Shapiro LG, Hanlon D, Martin RF, Brinkley JF, Poliakov AV, Ojemann GA, Corina DP. Evaluating spatial normalization methods for the human brain. In: Proceedings, 27th Annual Conference of the IEEE Engineering in Medicine and Biology Society (EMBS). Shanghai, China; 2005. p. 5331-5334. http://sigpubs.biostr.washington.edu/archive/00000185/.
53. Van Essen DC, Drury HA, Dickson J, Harwell J, Hanlon D, Anderson CH. An integrated software suite for surface-based analysis of cerebral cortex. J Am Med Ass 2001;8(5):443-459 http://stp.wustl.edu.
54. Re C, Brinkley J, Suciu D. A performant XQuery to SQL translator: University of Washington, Dept of Computer Science and Engineering Technical Report 2006. Report No.: 2006-06-02.http://sigpubs.biostr.washington.edu/archive/00000194/.
55. Re C. SilkRoute II - Efficient relational publishing to XML. 2006. http://silkroute.cs.washington.edu/.
56. Fernandez M, Kadiyska Y, Morishima A, Suciu D, Tan W. Silkroute: a framework for publishing relational data in XML. ACM Transactions on Database Technology 2002;27(4).
57. Tang Z, Kadiyska Y, Li H, Suciu D, Brinkley JF. Dynamic XML-based exchange of relational data: application to the Human Brain Project. In: Proceedings, Annual Fall Symposium of the American Medical Informatics Association. Washington, D.C.; 2003. p. 649-653. http://sigpubs.biostr.washington.edu/archive/00000141/.
58. Tang Z, Kadiyska Y, Suciu D, Brinkley JF. Results visualization in the XBrain XML interface to a relational database. In: Proceedings, MEDINFO. San Francisco, CA; 2004. p. 1878. http://sigpubs.biostr.washington.edu/archive/00000146/.
59. Structural Informatics Group. WIX: Web Interface for XQuery. 2007. http://sig.biostr.washington.edu/projects/wix/index.html.
60. Apache Software Foundation. Apache Tomcat. 2007. http://tomcat.apache.org/.
61. Kay M. SAXON XSLT and XQuery Processor. 2007. http://saxon.sourceforge.net/.
62. Halevy AY, Ives ZG, Madhavan J, Mork P, Suciu D, Tatarinov I. The Piazza peer data management system. IEEE Transactions on Knowledge and Data Engineering 2004;16:787-798.
63. Shaker R, Mork P, Brockenbrough JS, Donelson L, Tarczy-Hornoch P. The Biomediator system as a tool for integrating biologic databases on the web. In: Proc. Workshop on Information Integration on the Web, held in conjunction with VLDB; 2004. http://www.biomediator.org/publications/iiweb-paper-biomediator.pdf.
64. Wang K, Tarczy-Hornoch P, Shaker R, Mork P, Brinkley JF. BioMediator Data Integration: Beyond Genomics to Neuroscience Data. In: Proceedings, AMIA Fall Symposium; 2005. p. 779-783. http://sigpubs.biostr.washington.edu/archive/00000183/.
65. Jeng S, Wang K, Barbero J, Brinkley J, Tarczy-Hornoch P. A pilot bridging data integration and analytics: BioMediator and R. In: Proceedings American Medical Informatics Fall Symposium; 2005. p. 995.
66. Re C, Brinkley J, Hinshaw K, Suciu D. Distributed XQuery. In: Proceedings of the Workshop on Information Integration on the Web (IIWeb); 2004. p. 116-121. http://sigpubs.biostr.washington.edu/archive/00000157/.
67. Structural Informatics Group. DXQP - Distributed XQuery Processor. 2007. http://sig.biostr.washington.edu/projects/dxqp/.
68. Saxon. The XLST and XQuery Processor. 2006. http://saxon.sourceforge.net/.
69. Detwiler LT, Suciu D, Franklin JD, Moore EB, Lee ES, Corina D, Ojemann GA, Brinkley J. Lightweight data integration with XQuery. Submitted. 2007.
70. Li X. XGI: A graphical interface for XQuery creation and XML schema visualization [Masters]. Seattle: University of Washington; 2006.http://sigpubs.biostr.washington.edu/archive/00000198/
71. Wikipedia. Ajax. 2006. http://en.wikipedia.org/wiki/AJAX.
72. Corina DP, Steury KR, Mulligan KA, Hinshaw KP, Poliakov AP, Brinkley JF, Maravilla KR, Ojemann GA. A comparison of language cortex indentified by cortical stimulation mapping and fMRI techniques. In: Abstracts, Society for Neuroscience Annual Meeting. Miami; 1999. p. 654.2.
73. Lurito JT, Lowe MJ, Sartorius C, Mathews VP. Comparison of fMRI and intraoperative direct cortical stimulation in localization of receptive language areas. J Comput Assist Tomogr 2000;24(1):99-105 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10667669
74. Corina DP, Gibson EK, Martin RF, Poliakov A, Brinkley JF, Ojemann GA. Dissociation of action and object naming: evidence from cortical stimulation mapping. Hum Brain Mapp 2005;24(1):1-10 http://sigpubs.biostr.washington.edu/archive/00000165/.
75. Richards T, Oskin N, Panagiotides H, Webb S, Poliakov A, Aylward E, Dawson G. Face processing in autism: spatial-temporal localization of fusiform activation. In: Proceedings, 11th Annual Meeting of the Organization for Human Brain Mapping, Abstract #1162. Toronto, Ontario, Canada; 2005.
76. Johnson LC, Richards T, Poliakov A, Aylward E. Contrast masking and differential fMRI brain responses relative to a rest condition. In: Proceedings, 11th Annual Meeting of the Organization for Human Brain Mapping, Abstract #331. Toronto, Ontario, Canada; 2005. http://sigpubs.biostr.washington.edu/archive/00000195/.
77. Richards T, Oskin N, Panagiotides H, Webb S, Poliakov A, Kleinhans N, Aylward E, Dawson G. Face processing in autism: fMRI/EEG source localization correlations. In: 5th International Meeting for Autism Research (IMFAR). Montreal, Canada; 2006. p. 190.