MindSeer is a visualization tool for multi-modality neuroimaging data. It supports basic data management capabilities, visualization of 3D surfaces (SPM's output or OFF files), volumes (Analyze, NIFTI or Minc) and label sets.
For usage instructions and examples, see the MindSeer tutorials.
If you are using MindSeer in your work, please let us know and cite the following reference: Moore EB, Poliakov AV, Lincoln P, and Brinkley JF MindSeer: a portable and extensible tool for visualization of structural and functional neuroimaging data. BMC Bioinformatics 2007 8:389 doi:10.1186/1471-2105-8-389.
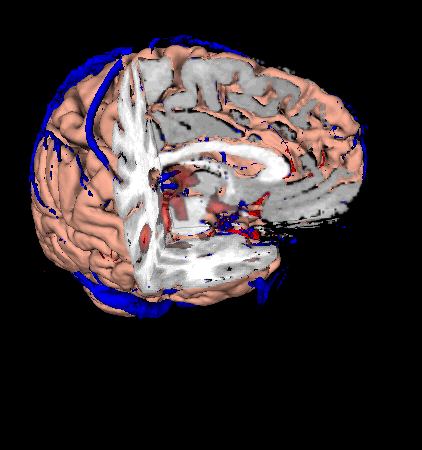
MindSeer has 2 different modes:
- Client/Server is designed to allow users to visualize data that is stored centrally and enhance collaboration.
- Standalone mode is available to view local data and is built for more performance than Client/Server
Both modes have the same interface and support the same features.
It has a modular architecture and is designed to be extensible. It is released under the GPL with some LGPL components (MatFile and NIFTI jar files).
Requirements
- Java 5.0 or above.
- Java Web Start.
- Java3D (installed automatically by Web Start).
Outside Uses
- Version 1.1 added support for atlases! Here is an example of BrainInfo's monkey atlas: Puzzle Atlas
- Project Wonderland at Sun has incorporated MindSeer into a demo of the "Science Playground": This is your brain in Wonderland