Seedpod
Data Management with Minimal Programming
Problem Statement
Biomedical researchers need data management tools that are more robust than spreadsheets. However, the rate at which biomedical informaticists have been developing laboratory information management systems (LIMS) can't keep up with the demand for LIMS and the rate that experiment protocols change.
Overview
Seedpod is a tool that allows informaticists and scientists to work together using Protégé to encode data concepts and relationships in a knowledge model, which is automatically transformed into a relational database schema. Seedpod also generates metadata for a transparent front end web-based information management application. Scientists use the web application for data management and access. No programming is required.
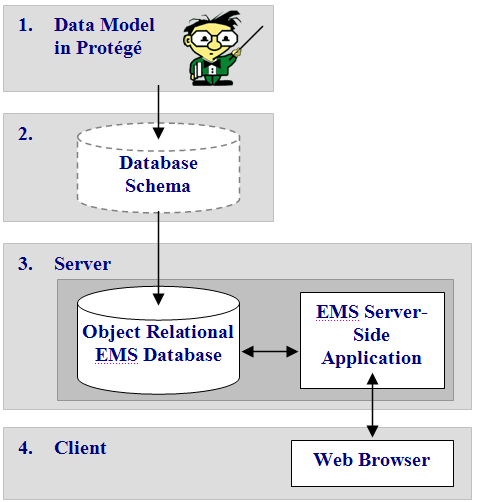
Figure 1 depicts the general system architecture. This architecture also allows the model to change independently without requiring informaticists to make changes to the web application. More information is available in this Seedpod overview presentation. We also have a simple demo Seedpod web application.
Seedpod is open source. Please contact us if you would like access to pre-release versions. Seedpod is written in Java and currently requires PostgreSQL and Apache Tomcat.
Supported by NIH Grants DC02310, LM07442, and RR025014.
Publications
- Li, Hao and Brinkley, James F and Gennari, John H (2004) Semi-automatic Database Design for Neuroscience Experiment Management Systems. In Proceedings, MedInfo 2004, page 1717, San Francisco, CA.
- Gennari, John H and Mork, Peter and Li, Hao (2005) Knowledge Transformations between Frame Systems and RDB Systems. In Proceedings, 3rd International Conference on Knowledge Capture (K-CAP'05), pages pp. 197-198, Banff, Alberta, Canada.
- Li, H., Gennari, JH, Brinkley, JF, Model-driven laboratory information management systems. Proeeedings AMIA Fall Symposium, 2006, pp 484-488.