Digital Anatomist Project Description
Goals
The long term goal of the Digital Anatomist project is an anatomy information system that is available from any desktop computer on the network. The development of this system is driven by the needs of students learning anatomy, but the system has now evolved to a state where it is used by clinicians as well. A user of the information system should be able to query the knowledge base for specific anatomic questions, to retrieve dynamically generated 3-D scenes illustrating answers to the query, and to use the retrieved information as the basis for queries of related databases and image repositories available on the network. Such an information system requires many modules including visual databases, 3-D modeling, real-time rendering, virtual reality and others. As in all our applications, our approach to meeting these requirements is an incremental one.

The current version of our anatomy information system has evolved from several previous versions. Although the current version does not satisfy all the long term requirements of an anatomy information system, it contains many components of that system, all within our overall conceptual framework.
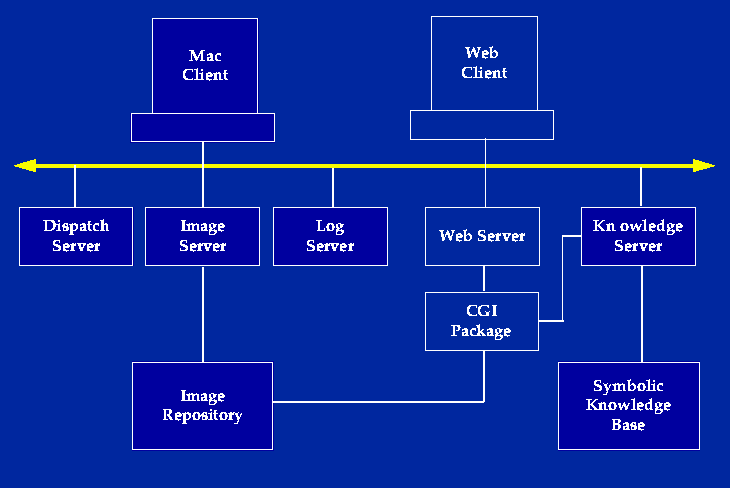
In the current system, the spatial database is an image repository that consists of sets of annotated images and animations packaged into interactive atlases representing different body regions. Most of the images and animations are renderings of 3-D models created by our in house authoring programs, although any annotated 2-D images may be utilized. The atlases may be offloaded to CD-ROM, where they are accessible by custom Mac and Windows clients. They also may be accessed directly by a custom Mac client that uses servers (the Dispatch Server, Image Server and Log Server), developed before the Web became popular [Brinkley 1993]. In the past few years most of the access has been via a Web client implemented by a set of CGI programs called the DA-CGI package [Bradley1995, Brinkley1997a]. The DA-CGI package also provides links to the symbolic knowledge server.
The construction of the animations and annotated images involves several image processing steps implemented by the authoring client programs. Ideally these steps should all be automatic once a 3-D anatomic model can be deformed to fit the data. However, since we have not solved this problem we use a more traditional approach, but with an eye toward integrating advances in knowledge-based segmentation from our own lab and from those of others.
The 3-D models are generated by a process of 3-D reconstruction from serial sections. The input is an image volume consisting of a set of serial sections. Two well-known examples of this kind of input are the Visible Human male and female, but clinical image volumes can also be used. The cross-section of each structure on each of these images is segmented, using a manual segmentation tool called Morpho. The resulting stack of contours is input to our locally-developed Skandha program. Skandha is then used to reconstruct the contours into a 3-D surface, to combine surfaces into 3-D models, and to render the models, either as static 2-D images or as Quicktime animations. In our production system most of these tasks are done manually (without the aid of shape knowledge), which is not a major burden since only one or two canonical models are being developed.
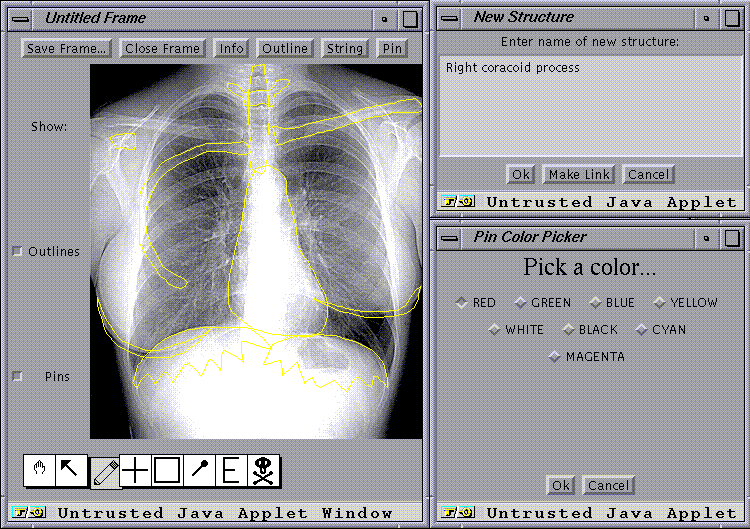
The 2-D images are annotated by a Java-based software tool we call Frame Builder, which allows the author to delineate regions on the images, and to label them either with the structure names or with commands to open other images. The annotations are saved in a separate file we call a frame. The combined animations and image-frame pairs are saved in a separate directory, one for each atlas.
The atlases are accessed over the network by means of the Atlas Web client, which is implemented by a set of C programs that constitute the DA-CGI package. For a given atlas the user navigates through the set of images by clicking on small image icons that retrieve the full image, or by searching for images that depict a structure. Once an annotated image is retrieved it can be examined in either browse mode or quiz mode.
Clicking here will take you to one of our atlas pages that shows browse mode for an annotated rendering of a 3-D model in our atlas of Thoracic Viscera. Simply click on regions of the image to get the names, then hit the back button to come back to this page.
Clicking here will take you to a 3-D image from our Brain atlas, that shows quiz mode. The computer asks you to click on a region that corresponds to the named structure.
In both these modes, state information is maintained by hidden form fields that are passed between the different DA-CGI programs.
Additional atlas client modes are described more fully in the online help page, and in other publications [Bradley1995,Brinkley1996]. These modes include a Pin Label mode, in which the names of all structures are arranged in the margin, and lines are drawn to the centers of the regions depicting the structures. Other modes include a table of contents mode, which allows the user to see the navigation hierarchy implicit in the frames, a knowledge base browse mode (not yet available in the production version), which displays the knowledge base hierarchy, and a search mode.
Search mode is entered either by typing a term name, or by clicking on a term in a knowledge base hierarchy. In either case, the search engine consults a separate relational database that contains the URLs for all images that contain the term. The user can then select from this list to retrieve the desired image. The search database is constructed by an ``atlas crawler'' program, similar to Web search engines such as the Web Crawler, that searches all atlases on the Web that are known to it.
Various versions of the anatomy information system have been in use since 1992, and evaluations have been in the context of gross anatomy and neuroanatomy education. However, we envision that this kind of information will be of use in many areas of clinical medicine, research and education. Evaluations have primarily addressed the following questions:
How useful and general is the software framework for entering and delivering image-based anatomical content, and
How is the atlas used?
We have also looked at the effect of network response time on the usefulness of the atlas [Dailey1993a,Dailey1994,Dailey1996].
The utility of the distributed framework for entering and delivering image-based anatomical content is evidenced by 1) the number of different interactive atlases that have been created by different authors, and 2) our ability to deliver the same information via CD-ROM as well as via net-based client programs.
The software framework has to date been used by our group to create atlases of the brain from a cadaver and from MRI of a living volunteer [Sundsten1992,Sundsten1994], an atlas of the thoracic viscera [Conley1994,Conley1994a] an atlas of the knee [Ratiu1994], and smaller atlases of the brainstem and chest radiology. The same material is available on videodisc [Conley1994a,Sundsten1992], CD-ROM [Conley1994,Sundsten1994,Ratiu1994], and on the Internet [Brinkley1993,Bradley1995].
The success and generality of the software framework has prompted the IAIMS program at the University of Washington to propose the Digital Anatomist anatomy information system as a tool that will be provided to faculty throughout the Health Sciences Center, and to universities in the four state area of the University of Washington's WAMI (Washington, Alaska, Montana, Idaho) regional medical education program. We expect that the authoring tools will soon be used by many authors to develop image-based atlases that can be linked over the Web. In fact, nothing requires the content to be medically-related, so it is likely that the tools will be of use in areas outside of medicine.
The usage of the various atlases has been assessed by our local experience, and by examination of on-line log files and comments.
Various versions of the atlas client have been in use for neuroanatomy and gross anatomy classes at the University of Washington since 1992, and by other institutions over the network since 1994. The Web version was introduced in June, 1995, and has since received six Web awards by outside rating organizations. Since Winter 1994 we have only used net-based access for our local courses because of the convenience to the authors in updating content.
During a one year period from June 1995 through June 1996 the total number of sites accessing the Web atlas was over 13,000 from 81 countries. Usage peaked during midterm and final exams, and was very low during breaks. The average Web daily requests was 4073, with a peak of about 25,000 requests on one day. On-line comments have been very favorable; the major requests are for more material, for other ways to navigate through the information, and for links to other parts of the anatomy curriculum.
The anatomy information system provides our most complete current example of an integrated system of the kind envisioned in our conceptual framework. For instance, the symbolic knowledge base is accessible both via the Knowledge Manager authoring program, and by the symbolic knowledge base server, which is in turn accessed by the atlas client. This means that as soon as the authors enter new information into the knowledge base, the information becomes available to the Web based term inspector and the knowledge browser in the atlas.
Similarly, as soon as authors create new annotated images, using Skandha and Frame Builder, the new annotated images become available throughout the world. This is one of the reasons why many remote users prefer the on-line atlas over the static CD-ROMS. (Another reason might be that the on-line atlases are currently free).
In addition to basic issues of 3-D modelling, much of the effort required to extend the anatomy information system will also involve integration, particularly between spatial and symbolic information. For example, once a complete set of anatomic terminology has been entered with the Knowledge Manager for all parts of the body, it will be necessary to ensure that all frames created by Frame Builder are labelled with terms from the Knowledge Base. For existing frames this can be done by an offline interactive lexical matching process. For newly created frames, however, the Java-based Frame builder will directly access the Symbolic Knowledge Server to display lists of terms that can be selected for annotating the images.
Once all the frames have been indexed by terms in the knowledge base, the atlas search engine can be made more intelligent, since it can use the semantic hierarchies in its searches (i.e., ``Find all frames that show any branch-of the ascending aorta''). The atlas client can also use the knowledge hierarchies to dynamically change the level of annotation detail: high levels, such as ``left ventricle'' or ``aorta'', for K-12 students, and lower levels, such as ``conus branch of right coronary artery'', for interventional radiologists and cardiac surgeons.
A second major integration step will enable the Web atlas client to call the Skandha program as a server, so that it can dynamically generate 3-D scenes that are annotated with structure names. The scenes can either be rendered on the server as static annotated images, or they can be sent to a VRML client for 3-D interactive viewing. Skandha can already be called as a server; however, the 3-D models need to be indexed by terms in the knowledge base, and the models need to be saved in a more accessible database. The repository manager being developed for the brain mapping project (see next section) should serve as a very useful management system for atlas models as well.
In the longer term, knowledge-based segmentation programs like Scanner will utilize combined 3-D spatial and symbolic knowledge to automatically instantiate patient-specific 3-D models from clinical image volumes, without the need for manual segmentation, reconstruction or image annotation. The models will then be accessed, by more advanced versions of the DA-CGI package, to generate on-line, realtime 3-D atlases, not just from canonical data like the Visible Human, but from any patient dataset.
The goals of the Digital Anatomist Project lead to many interesting informatics research issues, including image segmentation, graphics and visualization, artificial intelligence, and multimedia databases. Our progress in these areas is described in the research pages. As progress is made in these areas the results will be integrated with the Digital Anatomist information system, and with our other projects.