Ontology of Physics for Biology (OPB) Project
Background:
Central to the task of modern biology is representing the biological structures and the biological processes in which they participate at all levels of structural granularity (e.g., molecular, cellular, organ systems) across multiple scientific disciplines (e.g., molecular biology, systems physiology, organismal development). Until recent years such biological knowledge was available only in printed media organized and searched according to keywords (e.g., Google) and terminologies (e.g., PubMed). More recently, the bioinformatics community has developed a computational ontologies (see Open Biological Ontologies) have promised to capture and standardize the semantics of these resources enhance search capabilities and multidomain understanding. Whereas such ontologies have been very successful in the categorization and representation of biological structures (e.g., GeneOntology, Foundational Model of Anatomy), the representation of biological processes and the physics upon which they depend has received scant attention from the bioinformatical community.
Bioengineers and biophysicists have for years been representing and studying biological processes using physics-based mathematical models yet there is, as yet, no way to map between the sophisticated declarative knowledge of the biological structure and the sophisticated quantitative knowledge of biological processes. The need for such mappings has spurred the development of controlled terminologies such as the XML-based Systems Biology Markup Language and CellML. However, such terminologically-based languages are purpose-built for specific knowledge domains and do not leverage the knowledge represented in modern ontologies.
Consequently, we have initiated a two-pronged approach to provide strong semantic links between the ontologies of biological structure and the computational code of biosimulations. The first step is to develop an Ontology of Physics for Biology (OPB) as a reference ontology of those aspects of classical physics that are essential for representing, annotating and encoding quantitative models of biological processes. Just as the Foundational Model of Anatomy succeeds because of its strong theoretical underpinnings, the OPB is deeply based on theories of classical physics. Second, as described elsewhere on this site, we are implementing tools by which biosimulation models may be annotated, merged and encoded by reusing knowledge represented in the FMA and OPB.
Preliminary Work:
We have designed the OPB to be fully inclusive of a full range of biological processes and analytical approaches, yet we restrict this first version to so-called “lumped parameter” or discrete representations. Thus, for examples, we represent aortic blood pressure and the cellular concentration of glucose as single discrete properties (e.g., Paorta, Gcell) that have unique values at a moment in time for any instance of aortic blood and of cell. Such discretized, systems dynamical approach underlies the vast majority of biosimulation models in the literature and has a rich grounding in theories of engineering, physics and biology. the 3-dimensional spatial distributions of biological entities are reduced to and implicitly represented by scalar variables, parameters and equations (so-called “lumped-parameter” models).
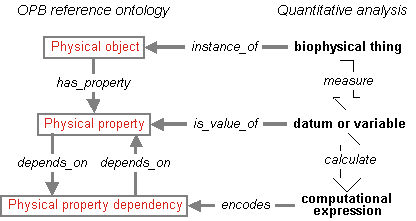
The overall schema of the OPB reference ontology and how it relates to quantitative analysis of biological systems is shown below: Physical objects (including body parts as represented in the FMA) have Physical properties (e.g., mass, acceleration) as attributes which are known by the laws of physics to depend upon each other by Physical property dependencies (e.g., Newton’s law; f = ma). These superclasses map readily to Quantitative analyses as encoded in a biosimulation model.
The OPB is intended to be multiscale and multidomain and therefore represents physical properties of all kinds relevant to biological investigation. Shown below is the OPB Physical property taxonomy which includes as major subclasses Displacement, Momentum, Flow, and Force, which themselves have subclasses for each (where appropriate) of seven “energy domains”: Fluid dynamics, Solid dynamics, Electricity, Chemical kinetics, Particle diffusion, Magnetism and Heat transfer.


Future goals:
We intend that subsequent OPB versions will build on the current version with support for other necessary modeling approaches such as: 1) continuum mechanics of spatially distributed biological processes such as the continuum mechanics of myocardial excitation-contraction and the reaction-diffusion kinetics of cellular signaling, 2) thermodynamics constraints on processes, and 3) statistical properties of sets or collections of instances.
We see the OPB as central to leveraging existing and evolving representations of biological knowledge and, in particular, for bridging the representational gap between the declarative representations of biological structure and the mathematical representations of biological processes.