Application Model Ontology (AMO) Project
Background:
Mathematical biosimulations model biological processes in a variety of computational environments (e.g., JSim, Fortran, MatLab..), yet the semantics of such models are implicit in the mathematics or, at best, embedded in in-line code annotations that are often opaque to anyone but model authors. Recognizing the need to reuse and integrate legacy and evolving biosimulation models, efforts are underway to annotate model libraries (e.g., BioModels, CellML repository) using XML-based languages. Such languages, however, lack the expressive power to identify critical model similarities and differences — particularly for multiscale biosimulation projects.
To establish annotation semantics of the required detail, we have proposed the Application Model Ontology (AMO; Pac Symp Biocomput. submitted 2008) as a light-weight semantic framework for building ApplModels — each of which is a symbolic representation that maps computational constructs (e.g., variables, equations) to classes in reference ontologies. Thus, ApplModels 1) annotate model variables by referencing Physical property (e.g., flow rate, tension) classes in the Ontology of Physics for Biology (OPB), 2) annotate variables as attributes of Anatomical entity classes (e.g., blood in aorta, cardiac myocyte) classes in the Foundational Model of Anatomy (FMA), and, 3) annotate model equations in terms of OPB Physical property dependency classes (e.g., Ohms Law, conservation of mass). In addition to reusing knowledge in available reference ontologies, ApplModels can be customized for idiosyncratic particular biomodelling tasks such as for pathological entities (e.g., trauma) and processes (e.g., hemorrhage).
Benefits to the biosimulation community include the ability to: 1) rigorously annotate and expose the semantic content of biosimulation models to support model reuse, 2) exploit strong semantics to symbolically modularize, merge, and augment multiscale models, 3) symbolically query and reason about simulation models to reveal their functional bases and input/output relations, 4) generate biosimulation code in a variety of languages according to the semantic structure of models, and 5) leverage available knowledge in Open Biological Ontologies (OBO).
Preliminary Work:
We have accomplished the following: 1) implemented AMO using the Protege ontology editor (see diagram below), 2) extended the Protege Prompt tab to derive a multiscale ApplModel that merges three independently-coded JSim biosimulation models: a closed-loop cardiovascular circulatory model, a baroreceptor model that relates heart rate and arteriolar resistance to aortic blood pressure, and a vascular smooth muscle model that relates calcium channel currents to arterial flow resistance, 3) demonstrated the generation of executable JSim code from an ontology-based model representation in our Chalkboard software (Pac Symp Biocomput. 2007; 12:16-27).
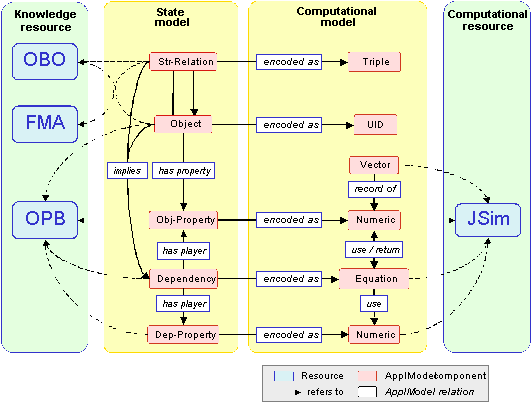
The diagram above illustrates parts of the schema for the Application Model Ontology. An ApplModel consists of a Physical model, that represents the structure and physics of the biological system, and a Computational model, whose computational constructs encode the components of the Physical model. This schema reuses and leverages declarative knowledge of various Knowledge resources (on the left) and maps to various Computational resources (on the right).
Future goals:
We envision that AMO-based ApplModels will substantially enhance biosimulation model reuse and integration for multiscale modeling tasks as diagrammed below.
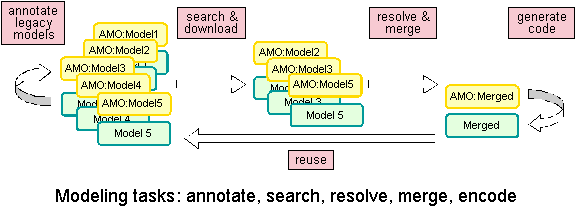
With these tasks in mind, we aim to develop tools to: 1) Annotate — implement an AMO-based Protege plug-in to read/parse legacy biosimulation code, to query the user for proper semantics of the code, and to output a ApplModel that fully annotates the code, 2) Search — develop methods to query ApplModel-annotated biosimulation model libraries to identify functionally relevant biosimulations. 3) Resolve & merge — extend the plug-in to input multiple ApplModels, resolve differences at the semantic level then output an ApplModel that represents a semantically merged version of the source models, 4) Generate code — migrate and encapsulate Chalkboard algorithms (Pac Symp Biocomput. 2007; 12:16-27) that will generate JSim code from ApplModel semantics.