The demo version of AnnoteImage that contacts the semantic web service embeds the following hardcoded SPARQL query in the Java code:
PREFIX fma_radlex: <http://bioontology.org/FMA_RadLex#>
PREFIX rdfs:<http://www.w3.org/2000/01/rdf-schema#>
PREFIX rdf:<http://www.w3.org/1999/02/22-rdf-syntax-ns#>
CONSTRUCT
{
?b fma_radlex:Preferred_Name ?name
}
WHERE
{?a fma_radlex:Preferred_Name "abdomen"^^<http://www.w3.org/2001/XMLSchema#string>
{?a fma_radlex:Has_regional_part ?b} UNION {?a fma_radlex:Has_constitutional_part ?b}
?b fma_radlex:Preferred_Name ?name .
}
You can see what this query does by cutting and pasting the text between the lines into a simple client that connects to the same semantic web service that the AnnoteImage program connects to. Before pasting the above query into the text box make sure you delete all the text that is already there, then hit the Submit Query button.
If you run the query as is you'll see that it generates the top level entities seen in the term search box in the AnnoteImage demo. Once the user opens up the hierarchy for that term in AnnoteImage a new query is sent to the web service, with the selected term subsitituted for "abdomen" in the above query. To see this try substituting "Abdomen proper" in the above query.
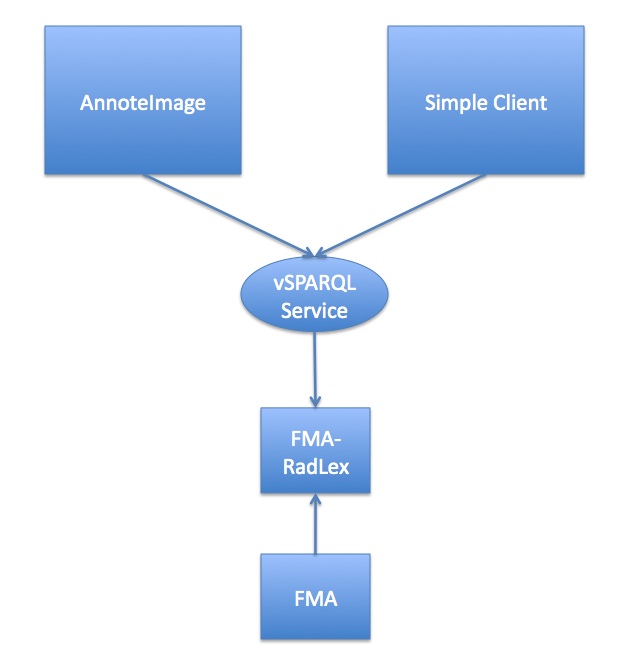
This figure shows the general architecture: We use a view query over the FMA to create FMA-RadLex, which is a materialized view of the FMA showing anatomy that is visible on a macroscopic image (leaving out entities like cells or molecules). The web service accesses this view, accepting vSPARQL queries and returning results to the client. In this demo there are two clients: AnnoteImage, and the Simple Client that lets you paste text into a text box.
